It’s exhausting to think about a time when “coronavirus” wasn’t a family phrase. But for a very long time, this household of viruses had merited little or no consideration. Believed to be ubiquitous amongst animals and avian species, the first coronavirus to infect and cause disease in humans was solely remoted and recognized within the Sixties.
Seven human coronaviruses have been identified since then.
Most cause solely comparatively minor well being issues: the common cold and seasonal respiratory infections that come round yearly. But the 2003 outbreak in China and different elements of Asia of severe acute respiratory syndrome (SARS), attributable to SARS-CoV (now renamed as SARS-CoV-1), propelled the virus onto the worldwide stage. Coronaviruses gained additional infamy when, in 2012, circumstances of the far more extreme Middle East respiratory syndrome (MERS) have been recognized in Saudi Arabia.
Both outbreaks have been comparatively contained. Not surprisingly, the priority over coronavirus ailments largely pale from the minds of abnormal individuals. The similar was true for virologists, who targeted their time and funding on extra urgent viruses. Then in late 2019 got here SARS-CoV-2, the causative agent of Covid-19.
Fortunately, some researchers had retained an curiosity in coronaviruses. After all, viruses can mutate and reappear, inflicting new outbreaks. One such cohort, ourselves amongst them, works on the University of the Western Cape in South Africa. Our laboratory had, amongst different issues, been learning a few of the structural proteins which might be the constructing blocks of coronaviruses. These proteins – named spike, nucleocapsid, membrane, and envelope proteins – have totally different roles, however are important to how coronaviruses reproduce, unfold and cause illness.
In our most recent paper, we examined what presumably units the human coronaviruses that cause SARS, MERS and Covid-19 aside from the opposite human coronaviruses that cause milder ailments like seasonal colds. The reply, we argue, lies with the envelope protein.
Shedding gentle on the E protein
The envelope protein is presumably probably the most enigmatic and least-studied within the coronavirus-suite, owing to its small measurement and the problem of learning it in laboratory settings. In May 2019, two of us printed a review paper on what was identified concerning the envelope protein on the time.
The paper has racked up almost 2,000 citations, most coming after the outbreak of Covid-19 – a testomony much less to our foresight than to the essential and beforehand understated function the envelope protein performs in human coronaviruses.
Even earlier than the Covid-19 outbreak, based mostly on what we had learnt from the SARS and MERS outbreaks, we have been satisfied that this protein – as soon as written off as a “minor component” of the virus – was key to the event of illness. It is essential, for example, within the remaining meeting of the virus, forming the envelope or wrapping that covers it when all its constituent elements come collectively.
It additionally performs a function within the virus’s budding, when it exits from the host cell; and within the course of often called pathogenesis, or the event and development of the an infection.
And it could maintain a clue to both the severity or relative mildness of the illness.
Our ongoing research is starting to recommend that the construction of the envelope protein could decide the severity of a coronavirus illness, or the distinction between a blocked nostril on the one hand, and collapsed lungs on the opposite.
The sting within the protein’s “tail”
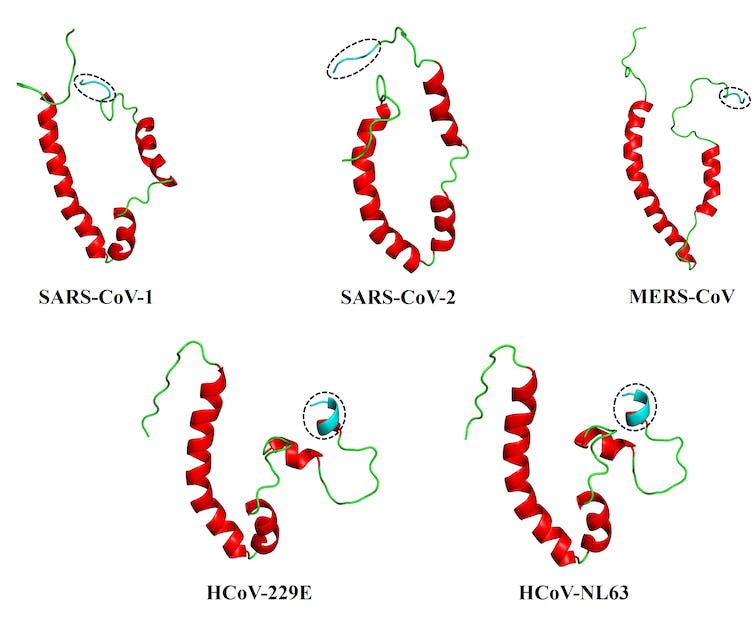
This led us to our most recent paper. We collaborated with structural bioinformatics professional Ruben Cloete, of the South African National Bioinformatics Institute on the University of the Western Cape, to develop full-length, 3D fashions of the envelope proteins of 5 human coronaviruses: SARS-CoV-1 and -2, and MERS-CoV (liable for the extreme SARS, Covid-19 and MERS ailments); and HCoV-229E and HCoV-NL63, liable for milder ailments. For this work, we relied on a modelling program often called MODELLER, permitting us to discover the proteins in some element.
Authors equipped
We then used a internet server, HADDOCK2.4, to simulate how the envelope protein interacts with the human PALS-1 protein – an interplay already proven to be essential with SARS-CoV-1. Each of the envelope proteins might bind to the PALS-1 protein, however the coronaviruses inflicting SARS, MERS and Covid-19 appeared to bind extra stably to PALS-1.
The solutions, we imagine, could lie within the conformation or form of what’s often called the PDZ-binding motif, or PBM, which sits on the tail-end of the envelope protein. This PBM – primarily a distinctive sequence on a protein – acts like a one-of-a-kind key to a very particular lock (often called the PDZ area) on a host cell protein. This ‘key’ permits the viral protein to work together with the host protein, making the illness worse.
We discovered that the extra versatile, prolonged coil of the PBM of the coronaviruses behind SARS, MERS and Covid-19 viruses could be what differentiates them from the extra inflexible PBM of the coronaviruses that cause milder ailments.
Inner workings
It is but too early to attract definitive conclusions, as these findings should be confirmed with extra research – within the laboratory and in residing organisms.
But it does shine some gentle on the internal workings of those coronaviruses and the still-enigmatic envelope protein. In so doing it might supply alternatives for the event of important life-saving remedies and vaccines.![]()
Dewald Schoeman, PhD Candidate, Molecular Biology and Virology, University of the Western Cape; Burtram C. Fielding, Dean Faculty of Natural Sciences and Professor, University of the Western Cape, and Ruben Cloete, Lecturer in Bioinformatics, University of the Western Cape
This article is republished from The Conversation beneath a Creative Commons license. Read the original article.